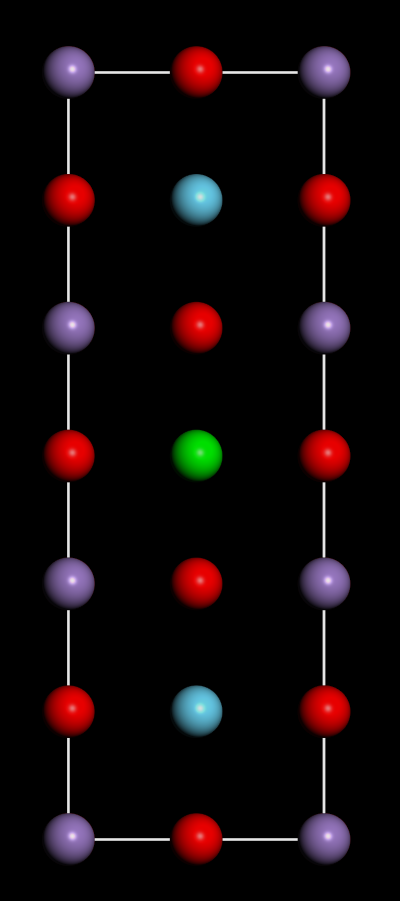
LSMO结构如下:
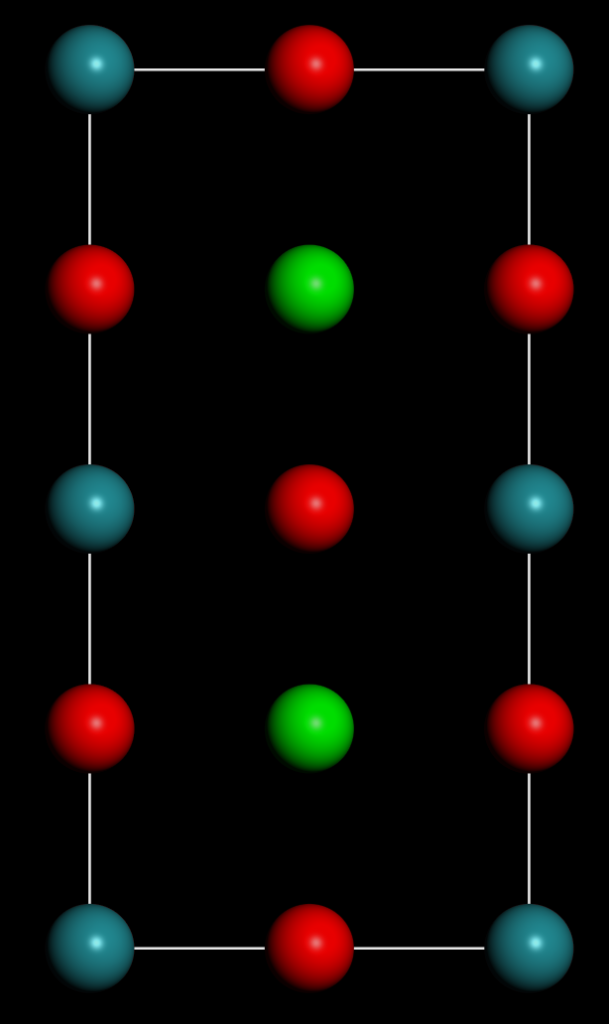
SRO结构如下,这里扩胞扩了两倍,不然后面会报错:
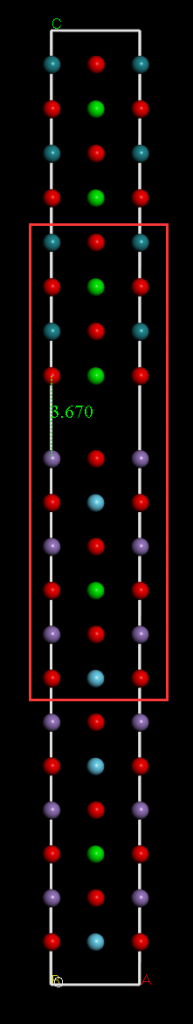
使用这两个结构构建散射区,注意要扩胞之后再建散射区,因为散射区包含了左右电极,如图所示,中间红框中为散射区,但是计算的时候要算整体。两层之间的距离应该扫描最低的能量来确定,这里我为了节约时间,随便设了一个数3.670埃
这几个结构的cif如下:
准备openmx的输入文件
左电极SRO:
# file names setting System.CurrrentDirectory ./ System.Name abc level.of.stdout 1 level.of.fileout 1 DATA.PATH /public/sourcecode/openmx_DATA19 NEGF.output_hks on NEGF.filename.hks SRO.hks Species.Number 3 <Definition.of.Atomic.Species O O6.0-s2p2d1 O_PBE19 Sr Sr10.0-s3p2d2 Sr_PBE19 Ru Ru7.0-s3p2d2 Ru_PBE19 Definition.of.Atomic.Species> Atoms.Number 5 Atoms.SpeciesAndCoordinates.Unit Ang <Atoms.SpeciesAndCoordinates 1 Sr 1.9700000 1.9700000 1.9700000 5.0 5.0 on 2 Ru 0.0000000 0.0000000 0.0000000 11 3 on 3 O 1.9700000 0.0000000 0.0000000 3.0 3.0 on 4 O 0.0000000 1.9700000 0.0000000 3.0 3.0 on 5 O 0.0000000 0.0000000 1.9700000 3.0 3.0 on Atoms.SpeciesAndCoordinates> Atoms.UnitVectors.Unit Ang <Atoms.UnitVectors 0.0000000 0.0000000 3.9400000 3.9400000 0.0000000 0.0000000 0.0000000 3.9400000 0.0000000 Atoms.UnitVectors> <Hubbard.U.values # eV O 1s 0.0 2s 0.0 1p 0.0 2p 0.0 1d 0.0 Sr 1s 0.0 2s 0.0 3s 0.0 1p 0.0 2p 0.0 1d 0.0 2d 0.0 Ru 1s 0.0 2s 0.0 3s 0.0 1p 0.0 2p 0.0 1d 3.0 2d 0.0 Hubbard.U.values> scf.XcType GGA-PBE scf.SpinPolarization off scf.ElectronicTemperature 300.0 scf.energycutoff 220.0 scf.maxIter 100 scf.EigenvalueSolver band scf.Kgrid 10 5 1 scf.Mixing.Type rmm-diisk scf.Init.Mixing.Weight 0.05 scf.Min.Mixing.Weight 0.01 scf.Max.Mixing.Weight 0.30 scf.Mixing.History 25 scf.Mixing.StartPulay 15 scf.criterion 1.0e-7 MD.Type nomd MD.maxIter 1 MD.TimeStep 1.0 MD.Opt.criterion 0.0003
注意这里修改了晶胞的方向,原来是:
Atoms.SpeciesAndCoordinates.Unit Ang <Atoms.SpeciesAndCoordinates 1 Sr 1.9700000 1.9700000 1.9700000 5.0 5.0 on 2 Ru 0.0000000 0.0000000 0.0000000 11 3 on 3 O 0.0000000 0.0000000 1.9700000 3.0 3.0 on 4 O 1.9700000 0.0000000 0.0000000 3.0 3.0 on 5 O 0.0000000 1.9700000 0.0000000 3.0 3.0 on Atoms.SpeciesAndCoordinates> Atoms.UnitVectors.Unit Ang <Atoms.UnitVectors 3.9400000 0.0000000 0.0000000 0.0000000 3.9400000 0.0000000 0.0000000 0.0000000 3.9400000 Atoms.UnitVectors>
改为:
Atoms.SpeciesAndCoordinates.Unit Ang <Atoms.SpeciesAndCoordinates 1 Sr 1.9700000 1.9700000 1.9700000 5.0 5.0 on 2 Ru 0.0000000 0.0000000 0.0000000 11 3 on 3 O 1.9700000 0.0000000 0.0000000 3.0 3.0 on 4 O 0.0000000 1.9700000 0.0000000 3.0 3.0 on 5 O 0.0000000 0.0000000 1.9700000 3.0 3.0 on Atoms.SpeciesAndCoordinates> Atoms.UnitVectors.Unit Ang <Atoms.UnitVectors 0.0000000 0.0000000 3.9400000 3.9400000 0.0000000 0.0000000 0.0000000 3.9400000 0.0000000 Atoms.UnitVectors>
右电极LSMO,注意这里同样改了晶胞的方向:
# # This was generated by OpenMX Viewer System.CurrrentDirectory ./ System.Name abc level.of.stdout 1 level.of.fileout 1 DATA.PATH /public/sourcecode/openmx_DATA19 NEGF.output_hks on NEGF.filename.hks LSMO.hks Species.Number 4 <Definition.of.Atomic.Species O O6.0-s2p2d1 O_PBE19 Mn Mn6.0-s3p2d1 Mn_PBE19 Sr Sr10.0-s3p2d2 Sr_PBE19 La La8.0-s3p2d2f1 La_PBE19 Definition.of.Atomic.Species> Atoms.Number 15 Atoms.SpeciesAndCoordinates.Unit Ang <Atoms.SpeciesAndCoordinates 1 O 0.0000000 1.9450000 0.0000000 3.0 3.0 on 2 O 0.0000000 0.0000000 1.9450000 3.0 3.0 on 3 O 1.9450389 0.0000000 0.0000000 3.0 3.0 on 4 La 1.9450389 1.9450000 1.9450000 7 4 on 5 Mn 0.0000000 0.0000000 0.0000000 11 4 on 6 O 3.8899611 1.9450000 0.0000000 3.0 3.0 on 7 O 3.8899611 0.0000000 1.9450000 3.0 3.0 on 8 O 5.8350000 0.0000000 0.0000000 3.0 3.0 on 9 Sr 5.8350000 1.9450000 1.9450000 5.0 5.0 on 10 Mn 3.8899611 0.0000000 0.0000000 11 4 on 11 O 7.7800389 1.9450000 0.0000000 3.0 3.0 on 12 O 7.7800389 0.0000000 1.9450000 3.0 3.0 on 13 O 9.7249611 0.0000000 0.0000000 3.0 3.0 on 14 La 9.7249611 1.9450000 1.9450000 7 4 on 15 Mn 7.7800389 0.0000000 0.0000000 7.5 7.5 on Atoms.SpeciesAndCoordinates> Atoms.UnitVectors.Unit Ang <Atoms.UnitVectors 0.0000000 0.0000000 11.6700000 3.8900000 0.0000000 0.0000000 0.0000000 3.8900000 0.0000000 Atoms.UnitVectors> <Hubbard.U.values # eV O 1s 0.0 2s 0.0 1p 0.0 2p 0.0 1d 0.0 Mn 1s 0.0 2s 0.0 3s 0.0 1p 0.0 2p 0.0 1d 4 Sr 1s 0.0 2s 0.0 3s 0.0 1p 0.0 2p 0.0 1d 0.0 2d 0.0 La 1s 0.0 2s 0.0 3s 0.0 1p 0.0 2p 0.0 1d 8.1 2d 0.0 1f 0.6 Hubbard.U.values> scf.XcType GGA-PBE scf.SpinPolarization off scf.ElectronicTemperature 300.0 scf.energycutoff 220.0 scf.maxIter 100 scf.EigenvalueSolver band scf.Kgrid 10 5 1 scf.Mixing.Type rmm-diisk scf.Init.Mixing.Weight 0.05 scf.Min.Mixing.Weight 0.01 scf.Max.Mixing.Weight 0.30 scf.Mixing.History 25 scf.Mixing.StartPulay 15 scf.criterion 1.0e-7 MD.Type nomd MD.maxIter 1 MD.TimeStep 1.0 MD.Opt.criterion 0.0003
提交任务后,分别得到SRO.hks和LSMO.hks
接下来计算散射区,openmx的输入文件如下:
# # This was generated by OpenMX Viewer System.CurrrentDirectory ./ System.Name abc level.of.stdout 1 level.of.fileout 1 DATA.PATH /public/sourcecode/openmx_DATA19 Species.Number 5 <Definition.of.Atomic.Species O O6.0-s2p2d1 O_PBE19 Mn Mn6.0-s3p2d1 Mn_PBE19 Sr Sr10.0-s3p2d2 Sr_PBE19 Ru Ru7.0-s3p2d2 Ru_PBE19 La La8.0-s3p2d2f1 La_PBE19 Definition.of.Atomic.Species> Atoms.Number 50 Atoms.SpeciesAndCoordinates.Unit Ang <Atoms.SpeciesAndCoordinates 1 O 23.2748991 1.9575000 0.0000000 3.0 3.0 on 2 O 23.2748991 0.0000000 1.9575000 3.0 3.0 on 3 O 1.8799250 0.0000000 0.0000000 3.0 3.0 on 4 La 1.8799250 1.9575000 1.9575000 7 4 on 5 Mn 23.2748991 0.0000000 0.0000000 11 4 on 6 O 3.8249226 1.9575000 0.0000000 3.0 3.0 on 7 O 3.8249226 0.0000000 1.9575000 3.0 3.0 on 8 O 5.7699203 0.0000000 0.0000000 3.0 3.0 on 9 Sr 5.7699203 1.9575000 1.9575000 5.0 5.0 on 10 Mn 3.8249226 0.0000000 0.0000000 11 4 on 11 O 7.7149179 1.9575000 0.0000000 3.0 3.0 on 12 O 7.7149179 0.0000000 1.9575000 3.0 3.0 on 13 O 9.6599156 0.0000000 0.0000000 3.0 3.0 on 14 La 9.6599156 1.9575000 1.9575000 7 4 on 15 Mn 7.7149179 0.0000000 0.0000000 11 4 on 16 O 11.6049132 1.9575000 0.0000000 3.0 3.0 on 17 O 11.6049132 0.0000000 1.9575000 3.0 3.0 on 18 O 13.5499109 0.0000000 0.0000000 3.0 3.0 on 19 La 13.5499109 1.9575000 1.9575000 7 4 on 20 Mn 11.6049132 0.0000000 0.0000000 11 4 on 21 O 15.4949085 1.9575000 0.0000000 3.0 3.0 on 22 O 15.4949085 0.0000000 1.9575000 3.0 3.0 on 23 O 17.4399061 0.0000000 0.0000000 3.0 3.0 on 24 Sr 17.4399061 1.9575000 1.9575000 5.0 5.0 on 25 Mn 15.4949085 0.0000000 0.0000000 11 4 on 26 O 19.3849038 1.9575000 0.0000000 3.0 3.0 on 27 O 19.3849038 0.0000000 1.9575000 3.0 3.0 on 28 O 21.3299015 0.0000000 0.0000000 3.0 3.0 on 29 La 21.3299015 1.9575000 1.9575000 7 4 on 30 Mn 19.3849038 0.0000000 0.0000000 11 4 on 31 Sr 26.9451684 1.9575000 1.9575000 5.0 5.0 on 32 Ru 40.7350877 0.0000000 0.0000000 11 3 on 33 O 26.9451684 0.0000000 0.0000000 3.0 3.0 on 34 O 40.7350877 1.9575000 0.0000000 3.0 3.0 on 35 O 40.7350877 0.0000000 1.9575000 3.0 3.0 on 36 Sr 30.8850246 1.9575000 1.9575000 5.0 5.0 on 37 Ru 28.9150965 0.0000000 0.0000000 11 3 on 38 O 30.8850246 0.0000000 0.0000000 3.0 3.0 on 39 O 28.9150965 1.9575000 0.0000000 3.0 3.0 on 40 O 28.9150965 0.0000000 1.9575000 3.0 3.0 on 41 Sr 34.8248808 1.9575000 1.9575000 5.0 5.0 on 42 Ru 32.8549527 0.0000000 0.0000000 11 3 on 43 O 34.8248808 0.0000000 0.0000000 3.0 3.0 on 44 O 32.8549527 1.9575000 0.0000000 3.0 3.0 on 45 O 32.8549527 0.0000000 1.9575000 3.0 3.0 on 46 Sr 38.7651595 1.9575000 1.9575000 5.0 5.0 on 47 Ru 36.7948089 0.0000000 0.0000000 11 3 on 48 O 38.7651595 0.0000000 0.0000000 3.0 3.0 on 49 O 36.7948089 1.9575000 0.0000000 3.0 3.0 on 50 O 36.7948089 0.0000000 1.9575000 3.0 3.0 on Atoms.SpeciesAndCoordinates> Atoms.UnitVectors.Unit Ang <Atoms.UnitVectors 0.0000000 0.0000000 42.2550000 3.9150000 0.0000000 0.0000000 0.0000000 3.9150000 0.0000000 Atoms.UnitVectors> <Hubbard.U.values # eV O 1s 0.0 2s 0.0 1p 0.0 2p 0.0 1d 0.0 Mn 1s 0.0 2s 0.0 3s 0.0 1p 0.0 2p 0.0 1d 4 Sr 1s 0.0 2s 0.0 3s 0.0 1p 0.0 2p 0.0 1d 0.0 2d 0.0 Ru 1s 0.0 2s 0.0 3s 0.0 1p 0.0 2p 0.0 1d 3.0 2d 0.0 La 1s 0.0 2s 0.0 3s 0.0 1p 0.0 2p 0.0 1d 8.1 2d 0.0 1f 0.6 Hubbard.U.values> scf.XcType GGA-PBE scf.SpinPolarization off scf.ElectronicTemperature 300.0 scf.energycutoff 220.0 scf.maxIter 100 scf.EigenvalueSolver NEGF scf.Kgrid 100 5 1 scf.Mixing.Type rmm-diisk scf.Init.Mixing.Weight 0.05 scf.Min.Mixing.Weight 0.01 scf.Max.Mixing.Weight 0.30 scf.Mixing.History 25 scf.Mixing.StartPulay 15 scf.criterion 1.0e-7 MD.Type nomd MD.maxIter 1 MD.TimeStep 1.0 MD.Opt.criterion 0.0003 NEGF.filename.hks.l LSMN.hks NEGF.filename.hks.r SRO.hks NEGF.Num.Poles 100 # defalut=150 NEGF.scf.Kgrid 30 1 # defalut=1 1 NEGF.SCF.Iter.Band 20 NEGF.Poisson.Solver FD # FD|FFT, default=FD NEGF.bias.voltage 0.0 NEGF.bias.neq.im.energy 0.01 # default=0.01 (eV) NEGF.bias.neq.energy.step 0.02 # default=0.02 (eV) NEGF.gate.voltage 1.0 # default=0.0 (in eV) NEGF.tran.Analysis on # default on NEGF.tran.energyrange -1 1 1.0e-3 # default=-10.0 10.0 1.0e-3 (eV) NEGF.tran.energydiv 3 # default=200 NEGF.tran.Kgrid 30 1 # default= 1 1
其中,NEGF.bias.voltage 0.0 代表电压是0V,如果要算输运性质,需要修改这个参数,重新计算openmx
计算完成后,得到以下文件:
如果出现以下错误:ERROR: PAOs of lead atoms can overlap only to the next nearest region. 这时需要扩胞,扩到2*2









No Comments
Leave a comment Cancel